这两天需要对预实验的脑电进行一个分类,在这里记录一下流程
脑电分析系列文章
mne官网
mne教程
随机森林分类
Python 多因素方差分析
文章目录
- 1. 脑电数据的处理
-
- 1.1 基本概念
- 1.2 实际处理
- 1.3 全部代码
- 2. 随机森林分类
-
- 1. label的制作
- 2. 使用随机森林进行分类
- 3. 全部代码
- 3. 显著性检验
- 4. 多文件测试
-
- 1. 文件选择
- 2. 精确度分析
- 3. anova分析
- 4. 可扩展性
-
- 1. 抽取代码
- 2. 有待扩展
1. 脑电数据的处理
1.1 基本概念
由于是刚刚学习的一些概念,这里就不做过多的解释贻笑大方了。就简单说一下自己的理解。
- raw :读取脑电的原始数据,里面重要的数据结构如下:
- info:记录一些备注信息,比如哪些是坏通道
- ch_name:采集数据用到的channel名字
- epoch:把原始的数据划分成段,方便后续的分析
- annotation/event: 每一段epoch的标签,两者区别具体看官方文档。不过用起来基本是一样的,并且有
mne.events_from_annotations和events_from_annotations可以相互转换。
了解完以上的概念之后,就可以进行实际的操作了。
1.2 实际处理
实际需求
- 由于采集数据的时候,没有进行annotation和event的标记,所以直接使用时间作为划分epoch的依据,这里采用30s作为一个epoch。
- 采集的是全64个通道,但是真正采集的就9个通道,需要对通道进行过滤
- 每20Min受试者报告一次或者两次本阶段的困倦程度,所以10 or 20min内的所有epoch都是一样的annotation
- 由于采集的时候电解液的风干,以及电极帽的接触不良,所以有的通道的数据可能不能用,应该进行过滤。
处理过程
1. 数据读取
import numpy as np
import mne
from mne.time_frequency import psd_welch
import os
################# 文件路径 ######################
filename = "D:/data/eeg/physionet-sleep-data/SC4001E0-PSG.edf"
filename2 = "D:/data/eeg/qyp.edf"
savepath = "result"
## 使用hxx作为训练数据, qyp作为测试数据
resultName = "features_test.npy"
# ############# 1. 读取文件 #################
raw = mne.io.read_raw(filename2, preload=True)
2. 数据预处理
数据预处理分成两步:
- 过滤不必要的频域
- 过滤不需要的通道
############# 2. 预处理 #####################
# 过滤高低频域,过滤防止0的干扰,同时减少数据量
raw.filter(l_freq=0.1, h_freq=40)
# 选择通道
alllist = ['Fpz', 'Fp1', 'Fp2', 'AF3', 'AF4', 'AF7', 'AF8', 'Fz', 'F1', 'F2', 'F3', 'F4', 'F5', 'F6', 'F7', 'F8', 'FCz', 'FC1', 'FC2', 'FC3', 'FC4', 'FC5', 'FC6', 'FT7', 'FT8', 'Cz', 'C1', 'C2', 'C3', 'C4', 'C5', 'C6', 'T7', 'T8', 'CP1', 'CP2', 'CP3', 'CP4', 'CP5', 'CP6', 'TP7', 'TP8', 'Pz', 'P3', 'P4', 'P5', 'P6', 'P7', 'P8', 'POz', 'PO3', 'PO4', 'PO5', 'PO6', 'PO7', 'PO8', 'Oz', 'O1', 'O2', 'ECG', 'HEOR', 'HEOL', 'VEOU', 'VEOL']
goodlist = ['FCz', 'Pz', 'Fpz', 'Cz', 'C1', 'C2','O1', 'O2', 'Oz',]
goodlist = set(goodlist)
badlist = []
for x in alllist:
if x not in goodlist:
badlist.append(x)
picks = mne.pick_channels(alllist, goodlist, badlist)
raw.plot(order=picks, n_channels=len(picks))
for x in badlist:
raw.info['bads'].append(x)
3. 按时间划分epoch
# 30s一个epoch
epochs = mne.make_fixed_length_epochs(raw, duration=30, preload=False)
4. 特征提取
def eeg_power_band(epochs):
"""脑电相对功率带特征提取
该函数接受一个""mne.Epochs"对象,
并基于与scikit-learn兼容的特定频带中的相对功率创建EEG特征。
Parameters
----------
epochs : Epochs
The data.
Returns
-------
X : numpy array of shape [n_samples, 5]
Transformed data.
"""
# 特定频带
FREQ_BANDS = {"delta": [0.5, 4.5],
"theta": [4.5, 8.5],
"alpha": [8.5, 11.5],
"sigma": [11.5, 15.5],
"beta": [15.5, 30]}
psds, freqs = psd_welch(epochs, picks='eeg', fmin=0.5, fmax=30.)
# 归一化 PSDs,这个数组中含有0元素,所以会出现问题,正确的解决方式,从epoch中去除或者从数组中去除
# psds = np.where(psds < 0.1, 0.1, psds)
# sm = np.sum(psds, axis=-1, keepdims=True)
# psds = numpy.divide(psds, sm)
psds /= np.sum(psds, axis=-1, keepdims=True)
X = []
for fmin, fmax in FREQ_BANDS.values():
psds_band = psds[:, :, (freqs >= fmin) & (freqs < fmax)].mean(axis=-1)
X.append(psds_band.reshape(len(psds), -1))
return np.concatenate(X, axis=1)
5. 保存文件
由于实验2个小时,所以会划分成240个epoch,但是有的可能会长,有的可能会短一个,长的应该直接舍去,因为这个是实验之后关闭设备采到的。
ans = eeg_power_band(epochs)
# 截取前240个数据
if ans.shape[0] > 240 :
ans = ans[:240]
print(ans.shape) ## hxx(240, 45), qyp (239, 45)
if not os.path.exists(savepath):
os.mkdir(savepath)
np.save(savepath + "/" + resultName, ans)
1.3 全部代码
"""
@author:fuzekun
@file:myFile.py
@time:2022/10/10
@description:
"""
import numpy as np
import mne
from mne.time_frequency import psd_welch
import os
################# 文件路径 ######################
filename = "D:/data/eeg/physionet-sleep-data/SC4001E0-PSG.edf"
filename2 = "D:/data/eeg/qyp.edf"
savepath = "result"
## 使用hxx作为训练数据, qyp作为测试数据
resultName = "features_test.npy"
# ############# 1. 读取文件 #################
raw = mne.io.read_raw(filename2, preload=True)
# ############ 2. 预处理 #####################
# # 过滤防止0的干扰
raw.filter(l_freq=0.1, h_freq=40)
# # 选择通道
alllist = ['Fpz', 'Fp1', 'Fp2', 'AF3', 'AF4', 'AF7', 'AF8', 'Fz', 'F1', 'F2', 'F3', 'F4', 'F5', 'F6', 'F7', 'F8', 'FCz', 'FC1', 'FC2', 'FC3', 'FC4', 'FC5', 'FC6', 'FT7', 'FT8', 'Cz', 'C1', 'C2', 'C3', 'C4', 'C5', 'C6', 'T7', 'T8', 'CP1', 'CP2', 'CP3', 'CP4', 'CP5', 'CP6', 'TP7', 'TP8', 'Pz', 'P3', 'P4', 'P5', 'P6', 'P7', 'P8', 'POz', 'PO3', 'PO4', 'PO5', 'PO6', 'PO7', 'PO8', 'Oz', 'O1', 'O2', 'ECG', 'HEOR', 'HEOL', 'VEOU', 'VEOL']
goodlist = ['FCz', 'Pz', 'Fpz', 'Cz', 'C1', 'C2','O1', 'O2', 'Oz',]
goodlist = set(goodlist)
badlist = []
for x in alllist:
if x not in goodlist:
badlist.append(x)
picks = mne.pick_channels(alllist, goodlist, badlist)
raw.plot(order=picks, n_channels=len(picks))
for x in badlist:
raw.info['bads'].append(x)
# ############## 2. 切分成epochs ################
epochs = mne.make_fixed_length_epochs(raw, duration=30, preload=False)
# ############# 3 特征提取 ##################
def eeg_power_band(epochs):
"""脑电相对功率带特征提取
该函数接受一个""mne.Epochs"对象,
并基于与scikit-learn兼容的特定频带中的相对功率创建EEG特征。
Parameters
----------
epochs : Epochs
The data.
Returns
-------
X : numpy array of shape [n_samples, 5]
Transformed data.
"""
# 特定频带
FREQ_BANDS = {"delta": [0.5, 4.5],
"theta": [4.5, 8.5],
"alpha": [8.5, 11.5],
"sigma": [11.5, 15.5],
"beta": [15.5, 30]}
psds, freqs = psd_welch(epochs, picks='eeg', fmin=0.5, fmax=30.)
# 归一化 PSDs,这个数组中含有0元素,所以会出现问题,正确的解决方式,从epoch中去除或者从数组中去除
# psds = np.where(psds < 0.1, 0.1, psds)
# sm = np.sum(psds, axis=-1, keepdims=True)
# psds = numpy.divide(psds, sm)
psds /= np.sum(psds, axis=-1, keepdims=True)
X = []
for fmin, fmax in FREQ_BANDS.values():
psds_band = psds[:, :, (freqs >= fmin) & (freqs < fmax)].mean(axis=-1)
X.append(psds_band.reshape(len(psds), -1))
return np.concatenate(X, axis=1)
ans = eeg_power_band(epochs)
# 截取前240个数据
if ans.shape[0] > 240 :
ans = ans[:240]
print(ans.shape) ## hxx(240, 45), qyp (239, 45)
if not os.path.exists(savepath):
os.mkdir(savepath)
np.save(savepath + "/" + resultName, ans)
2. 随机森林分类
1. label的制作
label制作分成两部分:
- 将原始的label从excel或者txt文件中提取出来
- 对原始label进行扩展,形成维度和特征相同的label列表
步骤1:
# 没有操作excel,直接复制粘贴的,应该采用excel的操作更加通用一些
"""
@author:fuzekun
@file:generateLabelFile.py
@time:2022/10/12
@description: 由原始标签生成每一个用户的id:label对应的set
"""
from collections import defaultdict
from json import dump, load
rawlabelFile = "result/rawLabel.json"
# 文件夹下面的名字
fileName = ["hxx.edf", "qyp.edf"]
# 对应的label
raws = []
raw1 = [(4, 5), 6, 7, 7, 6, 6] # hxx
raw2 = [4, 6, 7, (7, 6), (7, 5),5] #qup
raws.append(raw1)
raws.append(raw2)
ans = dict(zip(fileName, raws))
print(ans)
with open(rawlabelFile, 'w') as fp:
dump(ans, fp)
with open(rawlabelFile, 'r') as fp:
ans = load(fp)
print(ans)
将用户的文件名和label做一个映射:类似下面这种

步骤2:
def extractLabel(raw, n, spliteY, splitT, totalT):
"""
raw:原始标记
n: 特征的数量
spliteY: 划分的阈值
splitT: 划分epoch的时间 s
totalT: 总时常 h
"""
# 总共有多少段,然后每一个标签应该对应多少段
splitTime = totalT * 3600 // splitT // len(raw)
label = []
for i, x in enumerate(raw):
med = min(n, (i + 1) * splitTime)
mbg = i * splitTime
if (str.isdigit(str(x))):
x = 0 if x <= spliteY else 1 # [1,4]打成0,否则打成1
for _ in range(mbg, med):
label.append(x)
else: ## 一次回答两个数值的情况
x, y = x
x = 0 if x <= spliteY else 1
y = 0 if y <= spliteY else 1
for _ in range(mbg, mbg + splitTime // 2):
label.append(x)
for _ in range(mbg + splitTime // 2, med):
label.append(y)
return label
2. 使用随机森林进行分类
"""
@author:fuzekun
@file:myFile.py
@time:2022/10/10
@description:
"""
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score, classification_report
from sklearn.model_selection import train_test_split
X_file = "result/X.npy"
y_file = "result/y.npy"
X = np.load(X_file)
y = np.load(y_file)
Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y,test_size=0.3)
clf = RandomForestClassifier(max_depth=2,random_state=0)
clf.fit(Xtrain, Ytrain)
# print(clf.feature_importances_)
y_predict = clf.predict(Xtest)
acc = accuracy_score(Ytest, y_predict)
print("Accuracy score: {}".format(acc))
print(classification_report(Ytest, y_predict))

可以看到效果还是比较显著的。
3. 全部代码
- 整合数据
"""
@author:fuzekun
@file:extractFetures.py
@time:2022/10/12
@description: 特征提取
"""
import os
from json import load
import numpy as np
import mne
from mne.time_frequency import psd_welch
dataFilePath = "D:/data/eeg"
rawLabelFile = "result/rawLabel.json"
save_X = "D:/data/X.npy"
save_y = "D:/data/y.npy"
def extractFeture(dataFile:str, ch_name:list, spliteT:int, totalT:int):
"""
根据文件和chnnel提取特征
dataFile: 文件名
ch_name: 通道名称
spliteT:划分的时间s
totalT: 总时长h
return: 返回特征numpys数组
"""
# ############# 1. 读取文件 #################
raw = mne.io.read_raw(dataFile, preload=True)
# ############ 2. 预处理 #####################
# # 过滤防止0的干扰
raw.filter(l_freq=0.1, h_freq=40)
# # 选择通道
alllist = ['Fpz', 'Fp1', 'Fp2', 'AF3', 'AF4', 'AF7', 'AF8', 'Fz', 'F1', 'F2', 'F3', 'F4', 'F5', 'F6', 'F7', 'F8',
'FCz', 'FC1', 'FC2', 'FC3', 'FC4', 'FC5', 'FC6', 'FT7', 'FT8', 'Cz', 'C1', 'C2', 'C3', 'C4', 'C5', 'C6',
'T7', 'T8', 'CP1', 'CP2', 'CP3', 'CP4', 'CP5', 'CP6', 'TP7', 'TP8', 'Pz', 'P3', 'P4', 'P5', 'P6', 'P7',
'P8', 'POz', 'PO3', 'PO4', 'PO5', 'PO6', 'PO7', 'PO8', 'Oz', 'O1', 'O2', 'ECG', 'HEOR', 'HEOL', 'VEOU',
'VEOL']
goodlist = ch_name
goodlist = set(goodlist)
badlist = []
for x in alllist:
if x not in goodlist:
badlist.append(x)
# picks = mne.pick_channels(alllist, goodlist, badlist)
# raw.plot(order=picks, n_channels=len(picks))
for x in badlist:
raw.info['bads'].append(x)
# ############## 2. 切分成epochs ################
epochs = mne.make_fixed_length_epochs(raw, duration=spliteT, preload=False)
# ############# 3 特征提取 ##################
def eeg_power_band(epochs):
"""脑电相对功率带特征提取
该函数接受一个""mne.Epochs"对象,
并基于与scikit-learn兼容的特定频带中的相对功率创建EEG特征。
Parameters
----------
epochs : Epochs
The data.
Returns
-------
X : numpy array of shape [n_samples, 5]
Transformed data.
"""
# 特定频带
FREQ_BANDS = {"delta": [0.5, 4.5],
"theta": [4.5, 8.5],
"alpha": [8.5, 11.5],
"sigma": [11.5, 15.5],
"beta": [15.5, 30]}
psds, freqs = psd_welch(epochs, picks='eeg', fmin=0.5, fmax=30.)
# 归一化 PSDs,这个数组中含有0元素,所以会出现问题,正确的解决方式,从epoch中去除或者从数组中去除
# psds = np.where(psds < 0.1, 0.1, psds)
# sm = np.sum(psds, axis=-1, keepdims=True)
# psds = numpy.divide(psds, sm)
psds /= np.sum(psds, axis=-1, keepdims=True)
X = []
for fmin, fmax in FREQ_BANDS.values():
psds_band = psds[:, :, (freqs >= fmin) & (freqs < fmax)].mean(axis=-1)
X.append(psds_band.reshape(len(psds), -1))
return np.concatenate(X, axis=1)
ans = eeg_power_band(epochs)
n = totalT * 3600 // spliteT
# 截取前n个数据
if ans.shape[0] > n:
ans = ans[:n]
print(ans.shape) ## hxx(240, 45), qyp (239, 45)
return ans
def extractLabel(raw, n, spliteY, splitT, totalT):
"""
raw:原始标记
n: 特征的数量
spliteY: 划分的阈值
splitT: 划分epoch的时间 s
totalT: 总时常 h
"""
# 总共有多少段,然后每一个标签应该对应多少段
splitTime = totalT * 3600 // splitT // len(raw)
label = []
for i, x in enumerate(raw):
med = min(n, (i + 1) * splitTime)
mbg = i * splitTime
if (str.isdigit(str(x))):
x = 0 if x <= spliteY else 1 # [1,4]打成0,否则打成1
for _ in range(mbg, med):
label.append(x)
else: ## 一次回答两个数值的情况
x, y = x
x = 0 if x <= spliteY else 1
y = 0 if y <= spliteY else 1
for _ in range(mbg, mbg + splitTime // 2):
label.append(x)
for _ in range(mbg + splitTime // 2, med):
label.append(y)
return label
def extract(filePath, splitT, totalT, spliteY) : # 提取文件夹下的全部efg作为训练集,同时进行标签的填充
X = []
y = []
with open(rawLabelFile, 'r') as fp:
AllRawLabels = load(fp)
for fileName in os.listdir(filePath):
file = os.path.join(filePath, fileName)
if not os.path.isfile(file): continue
ch_name = ['FCz', 'Pz', 'Fpz', 'Cz', 'C1', 'C2','O1', 'O2', 'Oz',]
features = extractFeture(file, ch_name, splitT, totalT)
rawLabel = AllRawLabels[fileName]
labels = extractLabel(rawLabel, len(features), spliteY, splitT, totalT)
X.extend(list(features))
y.extend(labels)
return X, y
X, y = extract(dataFilePath, 30, 2)
X, y = np.array(X), np.array(y)
print(X.shape, y.shape)
np.save(save_X, X)
np.save(save_y, y)
- 随机森林
"""
@author:fuzekun
@file:myFile.py
@time:2022/10/10
@description:
"""
import numpy as np
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score, classification_report
from sklearn.model_selection import train_test_split
X_file = "result/X.npy"
y_file = "result/y.npy"
X = np.load(X_file)
y = np.load(y_file)
Xtrain, Xtest, Ytrain, Ytest = train_test_split(X,y,test_size=0.3)
clf = RandomForestClassifier(max_depth=2,random_state=0)
clf.fit(Xtrain, Ytrain)
# print(clf.feature_importances_)
y_predict = clf.predict(Xtest)
acc = accuracy_score(Ytest, y_predict)
print("Accuracy score: {}".format(acc))
print(classification_report(Ytest, y_predict))
3. 显著性检验
显著性检验分成四步
- 读取numpy数组
- 将numpy数组转化成pandas数组
- 使用函数进行anova分析,首先进行主观效应,其次进行多重比较检验
- 根据结果判断是否影响是否显著
"""
@author:fuzekun
@file:anova.py
@time:2022/10/12
@description: 方差分析
"""
import pandas as pd
import numpy as np
from statsmodels.formula.api import ols
from scipy import stats
import statsmodels.api as sm
import matplotlib.pyplot as plt
feature_name_list = []
for i in range(45) :
feature_name_list.append('f' + str(i))
# print(feature_name_list)
data_array = np.load("result/X.npy")
label_array = np.load("result/y.npy")
data_df = pd.DataFrame(data_array, columns=feature_name_list)
data_df['target'] = label_array
# print(data_df)
# print(label_df)
formula = 'target ~ '
for i, x in enumerate(feature_name_list):
if i != len(feature_name_list) - 1 :
formula += x + '+'
else :
formula += x
# print(data_df['f0'])
tired_anova = sm.stats.anova_lm(ols(formula,data = data_df).fit(),typ = 3)
print(tired_anova)
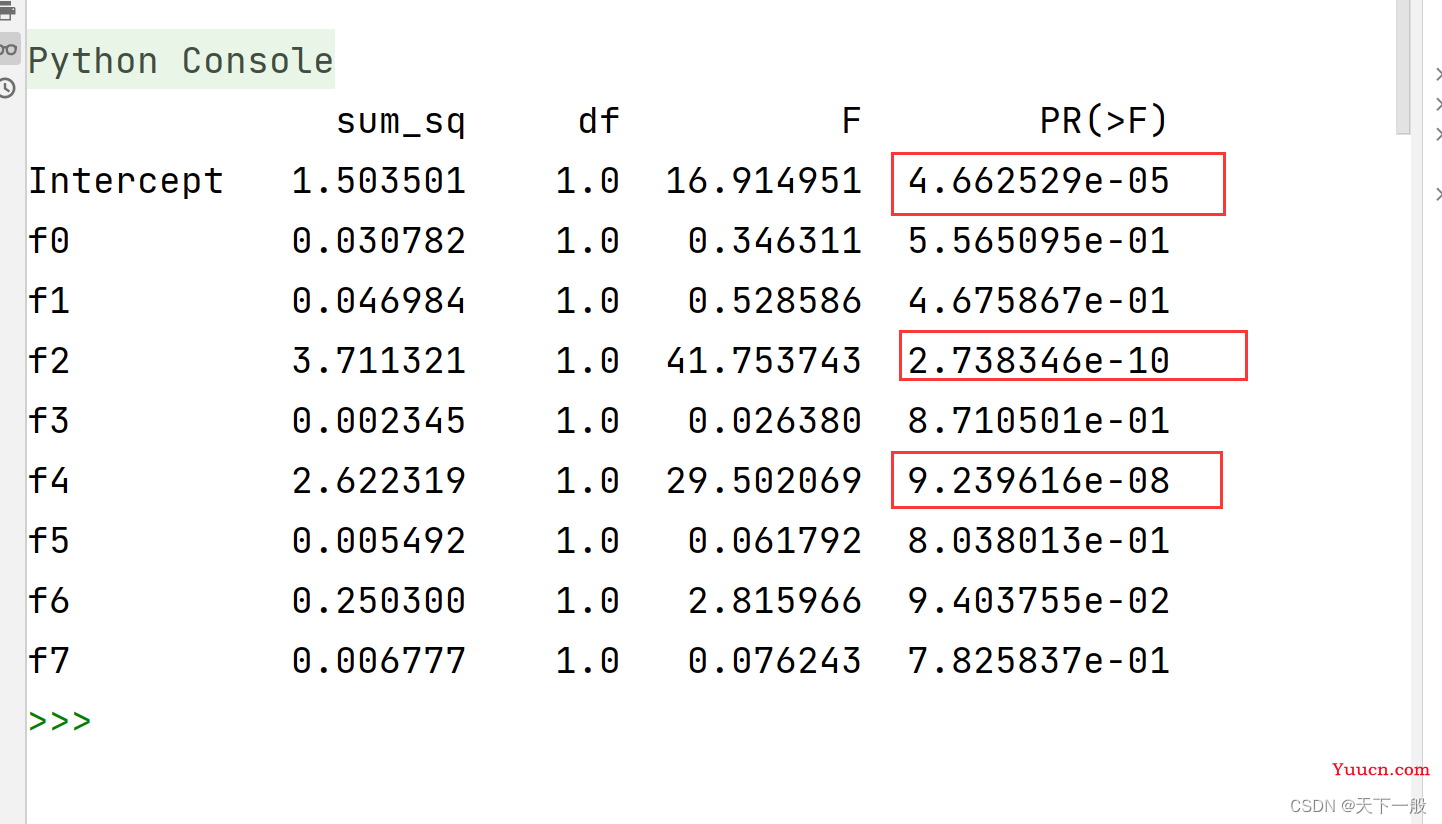
可以看到,上面的f0, f2, f4的远远小于了0.05, 也就是说拒绝了原假设,选择备择假设,可以说这些都是和target的相关性较强的点。
4. 多文件测试
1. 文件选择
使用了7个文件,选择了每一个文件的公共通道进行训练。部分脑电的图片如下所示:


2. 精确度分析
使用7个文件分类效果如下所示:精确度78%

3. anova分析
可以看到显著性较为明显

4. 可扩展性
1. 抽取代码
Json文件中存放了原始的标签。可以通过下面的代码生成,也可以直接进行编写。
"""
@author:fuzekun
@file:generateLabelFile.py
@time:2022/10/12
@description: 由原始标签生成每一个用户的id:label对应的set
"""
from collections import defaultdict
from json import dump, load
rawlabelFile = "result/rawLabel.json"
# 文件夹下面的名字
fileName = ["hxx.edf", "qyp.edf", "1_1.edf", "4_1.edf", "7_1.edf", "11_1.edf", "csk.edf"]
# 对应的label
raws = []
raw1 = [(4, 5), 6, 7, 7, 6, 6] # hxx
raw2 = [4, 6, 7, (7, 6), (7, 5),5] #qup
raw3 = [7, (9, 8), (5, 7), (5, 4), 4, 3] # 1_1
raw4 = [2, 5, 4, 5, 7, 8] # 4_1
raw5 = [7, 7, 8, 7, 6, 5] # 7_1
raw6 = [3, 5, 8, 6, 9, 5] # 11_1
raw7 = [3, 4, 5, 5, 6, 5] # csk
raws.append(raw1)
raws.append(raw2)
raws.append(raw3)
raws.append(raw4)
raws.append(raw5)
raws.append(raw6)
raws.append(raw7)
ans = dict(zip(fileName, raws))
print(ans)
with open(rawlabelFile, 'w') as fp:
dump(ans, fp)
with open(rawlabelFile, 'r') as fp:
ans = load(fp)
print(ans)
将label和特征的生成抽取成文件。
- 划分epoch的时间进行抽取
- 实验的时长,以及每一次询问的间隔。需要满足整数次
- 选择的通道进行抽取
"""
@author:fuzekun
@file:extractFetures.py
@time:2022/10/12
@description: 特征提取
"""
import os
from json import load
import numpy as np
import mne
from mne.time_frequency import psd_welch
### 原始的标签放在了rawLable.json文件中,可以进行编写,也可以直接修改generateLableFile生成。
dataFilePath = "D:/data/eeg"
rawLabelFile = "result/rawLabel.json"
save_X = "result/X.npy"
save_y = "result/y.npy"
ch_name = ['FCz', 'Pz', 'Fpz', 'Cz', 'C1', 'C2', ]
def extractFeture(dataFile:str, ch_name:list, spliteT:int, totalT:int):
"""
根据文件和chnnel提取特征
dataFile: 文件名
ch_name: 通道名称
spliteT:划分的时间s
totalT: 总时长h
return: 返回特征numpys数组
totalT * 3600 / spliteT一定要能整除。
"""
# ############# 1. 读取文件 #################
raw = mne.io.read_raw(dataFile, preload=True)
# ############ 2. 预处理 #####################
# # 过滤防止0的干扰
raw.filter(l_freq=0.1, h_freq=40)
# # 选择通道
alllist = ['Fpz', 'Fp1', 'Fp2', 'AF3', 'AF4', 'AF7', 'AF8', 'Fz', 'F1', 'F2', 'F3', 'F4', 'F5', 'F6', 'F7', 'F8',
'FCz', 'FC1', 'FC2', 'FC3', 'FC4', 'FC5', 'FC6', 'FT7', 'FT8', 'Cz', 'C1', 'C2', 'C3', 'C4', 'C5', 'C6',
'T7', 'T8', 'CP1', 'CP2', 'CP3', 'CP4', 'CP5', 'CP6', 'TP7', 'TP8', 'Pz', 'P3', 'P4', 'P5', 'P6', 'P7',
'P8', 'POz', 'PO3', 'PO4', 'PO5', 'PO6', 'PO7', 'PO8', 'Oz', 'O1', 'O2', 'ECG', 'HEOR', 'HEOL', 'VEOU',
'VEOL']
goodlist = ch_name
goodlist = set(goodlist)
badlist = []
for x in alllist:
if x not in goodlist:
badlist.append(x)
# picks = mne.pick_channels(alllist, goodlist, badlist)
# raw.plot(order=picks, n_channels=len(picks))
for x in badlist:
raw.info['bads'].append(x)
# ############## 2. 切分成epochs ################
epochs = mne.make_fixed_length_epochs(raw, duration=spliteT, preload=False)
# ############# 3 特征提取 ##################
def eeg_power_band(epochs):
"""脑电相对功率带特征提取
该函数接受一个""mne.Epochs"对象,
并基于与scikit-learn兼容的特定频带中的相对功率创建EEG特征。
Parameters
----------
epochs : Epochs
The data.
Returns
-------
X : numpy array of shape [n_samples, 5]
Transformed data.
"""
# 特定频带
FREQ_BANDS = {"delta": [0.5, 4.5],
"theta": [4.5, 8.5],
"alpha": [8.5, 11.5],
"sigma": [11.5, 15.5],
"beta": [15.5, 30]}
psds, freqs = psd_welch(epochs, picks='eeg', fmin=0.5, fmax=30.)
# 归一化 PSDs,这个数组中含有0元素,所以会出现问题,正确的解决方式,从epoch中去除或者从数组中去除
# psds = np.where(psds < 0.1, 0.1, psds)
# sm = np.sum(psds, axis=-1, keepdims=True)
# psds = numpy.divide(psds, sm)
psds /= np.sum(psds, axis=-1, keepdims=True)
X = []
for fmin, fmax in FREQ_BANDS.values():
psds_band = psds[:, :, (freqs >= fmin) & (freqs < fmax)].mean(axis=-1)
X.append(psds_band.reshape(len(psds), -1))
return np.concatenate(X, axis=1)
ans = eeg_power_band(epochs)
n = totalT * 3600 // spliteT
# 截取前n个数据
if ans.shape[0] > n:
ans = ans[:n]
print(ans.shape)
return ans
def extractLabel(raw, n, spliteY, splitT, totalT):
"""
raw:原始标记
n: 特征的数量
spliteY: 划分的阈值
splitT: 划分epoch的时间 s
totalT: 总时常 h
"""
# 总共有多少段,然后每一个标签应该对应多少段
# 注意这个一定要能整除,否则出现标签数量过少的情况
splitTime = totalT * 3600 // splitT // len(raw)
label = []
for i, x in enumerate(raw):
med = min(n, (i + 1) * splitTime)
mbg = i * splitTime
if (str.isdigit(str(x))):
x = 0 if x <= spliteY else 1 # [1,4]打成0,否则打成1
for _ in range(mbg, med):
label.append(x)
else: ## 一次回答两个数值的情况
x, y = x
x = 0 if x <= spliteY else 1
y = 0 if y <= spliteY else 1
for _ in range(mbg, mbg + splitTime // 2):
label.append(x)
for _ in range(mbg + splitTime // 2, med):
label.append(y)
return label
def extract(filePath, splitT, totalT, spliteY, ch_name) : # 提取文件夹下的全部efg作为训练集,同时进行标签的填充
X = []
y = []
with open(rawLabelFile, 'r') as fp:
AllRawLabels = load(fp)
for fileName in os.listdir(filePath):
print('##################' + fileName + '###################')
file = os.path.join(filePath, fileName)
if not os.path.isfile(file): continue
features = extractFeture(file, ch_name, splitT, totalT)
rawLabel = AllRawLabels[fileName]
labels = extractLabel(rawLabel, len(features), spliteY, splitT, totalT)
print(len(features), len(labels))
X.extend(list(features))
y.extend(labels)
return X, y
X, y = extract(dataFilePath, 30, 2, 4, ch_name)
X, y = np.array(X), np.array(y)
print(X.shape, y.shape)
np.save(save_X, X)
np.save(save_y, y)
2. 有待扩展
- 应该从excel文件中直接读取,但是没有这样做,还需要手动写。
- 实验如果有中断,每一个文件的时常会变化,单是
extract()函数默认每一个文件的时长和划分的间隔都是一样的。可以修改extract函数,将每一个文件的时长形成列表。然后分别extractLable和extractFeatur